23 void run_validation_test(){
34 float scale_factor = 1.0f;
44 int N_ind = scale_factor*pow(10.f,4);
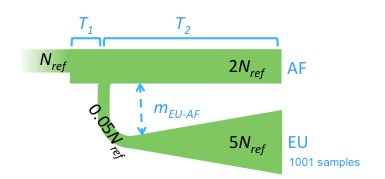
45 dem_const pop0(N_ind);
47 dem_pop_const gen0(pop0,pop1,1);
48 dem_const pop0_final(2*N_ind);
49 dem_pop_const gen1(pop0_final,pop1,1);
50 init_expansion gen_0_1(gen0,gen1,1);
51 exp_growth pop1_gen100((log(100.f)/(scale_factor*900.f)),0.05*N_ind,scale_factor*100);
52 dem_pop_const_exp gen100(pop0_final,pop1_gen100,1);
55 mig_const no_mig_pop0;
56 mig_dir no_pop1_gen0(0.f,1,1,no_mig_pop0);
57 mig_split create_pop1(1.f,0,1,no_pop1_gen0);
58 split_pop0 migration_split(no_mig_pop0,create_pop1,scale_factor*100);
59 float mig = 1.f/(2.f*N_ind);
68 int sample_size = 1001;
73 cudaEvent_t start, stop;
75 cudaEventCreate(&start);
76 cudaEventCreate(&stop);
78 float avg_num_mutations = 0;
79 float avg_num_mutations_sim = 0;
80 std::vector<std::vector<float> > results(num_iter);
81 for(
int j = 0; j < num_iter; j++){ results[j].reserve(sample_size); }
83 for(
int j = 0; j < num_iter; j++){
84 if(j == num_iter/2){ cudaEventRecord(start, 0); }
91 avg_num_mutations += ((float)my_spectra.
num_mutations)/num_iter;
93 for(
int i = 0; i < sample_size; i++){ results[j][i] = my_spectra.
frequency_spectrum[i]; }
97 cudaEventRecord(stop, 0);
98 cudaEventSynchronize(stop);
99 cudaEventElapsedTime(&elapsedTime, start, stop);
100 cudaEventDestroy(start);
101 cudaEventDestroy(stop);
104 std::cout<<
"SFS :"<<std::endl<<
"allele count\tavg# mutations\tstandard dev\tcoeff of variation (aka relative standard deviation)"<< std::endl;
105 for(
int i = 1; i < sample_size; i++){
109 for(
int j = 0; j < num_iter; j++){ num_mutations = b.
num_sites() - results[j][0]; avg += results[j][i]/(num_iter*num_mutations); }
110 for(
int j = 0; j < num_iter; j++){ num_mutations = b.
num_sites() - results[j][0]; std += 1.0/(num_iter-1)*pow(results[j][i]/num_mutations-avg,2); }
112 std::cout<<i<<
"\t"<<avg<<
"\t"<<std<<
"\t"<<(std/avg)<<std::endl;
116 std::cout<<
"\naverage number of mutations in simulation: "<<avg_num_mutations_sim<<
"\naverage number of mutations in SFS: "<<avg_num_mutations<<
"\ntime elapsed (ms): "<< 2*elapsedTime/num_iter<<std::endl;
123 int main(
int argc,
char **argv) { run_validation_test(); }
functor: models selection coefficient s as a constant across populations and over time ...
float * frequency_spectrum
site frequency spectrum data structure
int seed2
random number seed 2 of 2
functor: migration function changes from m1 to m2 at generation inflection_point
functor: turns sampling and preserving off (for every generation except the final one which is always...
control and output data structure for GO_Fish simulation
int compact_interval
how often to compact the simulation and remove fixed or lost mutations
functor: single, constant population size (N individuals) across populations and over time ...
functor: migration flows at rate m from pop1 to pop2 and function rest for all other migration rates ...
site frequency spectrum data structure (at the moment, functions only generate SFS for a single popul...
functor: one population, pop, has a different, demography function, d_pop, all others have function...
int num_populations
number of populations in simulation
proto-API for building site frequency spectra (contains the titular namespace Spectrum) ...
float num_mutations
number of segregating mutations in SFS
int maximal_num_mutations()
returns number of reported mutations in the final time sample (maximal number of stored mutations in ...
void site_frequency_spectrum(SFS &mySFS, const GO_Fish::allele_trajectories &all_results, const int sample_index, const int population_index, const int sample_size, int cuda_device=-1)
create a single-population SFS of size sample_size from a single time point sample_index in a single ...
int seed1
random number seed 1 of 2
functor: models parameter p as a constant across populations and over time
functor: migration flows at rate m from pop i to pop j =/= i and 1-(num_pop-1)*m for i == j ...
GO Fish Simulation API (contains namespaces GO_Fish and Sim_Model)
functor: models exponential growth of population size (individuals) over time
__host__ void run_sim(allele_trajectories &all_results, const Functor_mutation mu_rate, const Functor_demography demography, const Functor_migration mig_prop, const Functor_selection sel_coeff, const Functor_inbreeding FI, const Functor_dominance dominance, const Functor_preserve preserve_mutations, const Functor_timesample take_sample)
runs a single-locus Wright-Fisher simulation specified by the given simulation functions and sim_cons...
float num_sites
number of sites in simulation
int num_sites()
returns the number of sites in the simulation
sim_constants sim_input_constants
constants for initializing the next simulation
sim_constants last_run_constants()
returns sim_constants of the simulation currently held by allele_trajectories
functor: demography function changes from d1 to d2 at generation inflection_point ...
int num_generations
number of generations in simulation
4 from numpy
import array
7 def prior_onegrow_mig((nu1F, nu2B, nu2F, m, Tp, T), (n1,n2), pts):
9 Model with growth, split, bottleneck in pop2, exp recovery, migration 11 nu1F: The ancestral population size after growth. (Its initial size is 13 nu2B: The bottleneck size for pop2 14 nu2F: The final size for pop2 15 m: The scaled migration rate 16 Tp: The scaled time between ancestral population growth and the split. 17 T: The time between the split and present 19 n1,n2: Size of fs to generate. 20 pts: Number of points to use in grid for evaluation. 23 xx = yy = dadi.Numerics.default_grid(pts)
26 phi = dadi.PhiManip.phi_1D(xx, gamma=gamma_model)
28 phi = dadi.Integration.one_pop(phi, xx, Tp, nu=nu1F, gamma=gamma_model)
31 phi = dadi.PhiManip.phi_1D_to_2D(xx, phi)
34 nu2_func =
lambda t: nu2B*(nu2F/nu2B)**(t/T)
35 phi = dadi.Integration.two_pops(phi, xx, T, nu1=nu1F, nu2=nu2_func, m12=m, m21=m, gamma1=gamma_model, gamma2=gamma_model)
38 sfs = dadi.Spectrum.from_phi(phi, (n1,n2), (xx,yy))
41 def runModel(nu1F, nu2B, nu2F, m, Tp, T):
44 print 'sample sizes:', ns
49 func = prior_onegrow_mig
50 params = array([nu1F, nu2B, nu2F, m, Tp, T])
53 func_ex = dadi.Numerics.make_extrap_func(func)
56 model = func_ex(params, ns, pts_l)
57 model = model.marginalize((0,))
63 model = runModel(2, 0.05, 5, 1, 0.005, 0.045)
66 print "total runtime (s): " + str(time2-time1)
67 model = model/model.S()
69 for i
in range(0,len(model)):
Below is the makefile to create executable GOFish from examples/example_dadi/run.cu, each line is documented by the top part of the makefile:
Tip: The makefile below compiles machine code explicitly for generation 3.0 and 5.2 GPUs and uses just in time (JIT) compilation for everything else (lowest GPU generation which works for 3P is 3.0). Compilation (and program execution) will be faster if compiling for your specific GPU.
e.g. if running a Tesla K20 or Tesla K40, then the corresponding GPU generation is 3.5: all the current --generate-code arch=##,code=## flags can be deleted and replaced with --generate-code arch=compute_35,code=sm_35.
1 # Description of Mac/Linux/Unix Makefile for example_dadi.
3 #############################
4 # build_path := Where to build program and put executable (note: folder must already exist)
5 # api_path_source := Location of API source folder
6 # api_path_include := Location of API include folder
7 # EXEC_FILE := Name of executable
9 # NVCC := Compiler path, in this case nvcc is in $PATH
10 # CFLAGS := Compiler Flags: optimize most, fast math, add API include folder to include search path, equivalent to --relocatable-device-code=true --compile
11 # CODE := GPU types for which to build explicitly (I have a NVIDIA GTX 780M and 980) https://developer.nvidia.com/cuda-gpus, creates machine code for code=sm_30 (780) and code=sm_52 (980) and virtual architectures for all other generations which can be compiled JIT - code=compute_30 for generations between (3.0,5.0) and code=compute_50 for generations (5.0 and up) http://docs.nvidia.com/cuda/cuda-compiler-driver-nvcc/index.html#options-for-steering-gpu-code-generation
13 # object_names := Objects required for executable
14 # objects := Prepends build path to object names
16 # all := Command 'make' or 'make all' builds executable file $(EXEC_FILE) in location $(build_path)/
18 # ##### Object Dependencies Lists #####
19 # If one of the files on the right hand side of the : changes, the corresponding object must be recompiled.
20 # This is a developer's makefile - it assumes there will be changes to the GOFish API files, if not
21 # all the non-*.cu (non source file) dependencies are unnecessary.
22 # ##### End Object Dependencies Lists #####
24 # $(objects) := Make target all objects
25 # Compile source code into objects, $< := dependencies from Object Dependencies Lists, $@ := object in $(objects)
27 # $(build_path)/$(EXEC_FILE) := Make target executable EXEC_FILE which depends on all objects
28 # Link objects into executable EXEC_FILE, $@ := $(build_path)/$(EXEC_FILE)
30 # .PHONY := Defines 'all' and 'clean' as not true targets (i.e. don't remake executable if can't find files called 'all' or 'clean')
32 # clean := Action to perform for command 'make clean'
33 # Remove all objects and EXEC_FILE from build_path
34 #############################
36 build_path = ../example_dadi
37 api_path_source = ../../3P/_internal
38 api_path_include = ../../3P
42 CFLAGS = -O3 --use_fast_math -I $(api_path_include)/ -dc
43 CODE = --generate-code arch=compute_30,code=sm_30 --generate-code arch=compute_52,code=sm_52 --generate-code arch=compute_30,code=compute_30 --generate-code arch=compute_52,code=compute_52
45 object_names = run.o spectrum.o shared.o go_fish_impl.o
46 objects = $(addprefix $(build_path)/,$(object_names))
48 all:$(build_path)/$(EXEC_FILE)
50 ##### OBJECT DEPENDENCIES #####
51 $(build_path)/run.o: run.cu $(api_path_source)/shared.cuh $(api_path_source)/go_fish_impl.cuh $(api_path_include)/go_fish_data_struct.h $(api_path_include)/go_fish.cuh $(api_path_include)/spectrum.h
52 $(build_path)/spectrum.o: $(api_path_source)/spectrum.cu $(api_path_include)/spectrum.h $(api_path_source)/shared.cuh $(api_path_include)/go_fish_data_struct.h
53 $(build_path)/shared.o: $(api_path_source)/shared.cu $(api_path_source)/shared.cuh
54 $(build_path)/go_fish_impl.o: $(api_path_source)/go_fish_impl.cu $(api_path_source)/go_fish_impl.cuh $(api_path_source)/shared.cuh
55 ##### END OBJECT DEPENDENCIES #####
58 $(NVCC) $(CODE) $(CFLAGS) $< -o $@
60 $(build_path)/$(EXEC_FILE): $(objects)
62 $(NVCC) $(CODE) $(objects) -o $@
67 rm -f $(objects) $(build_path)/$(EXEC_FILE)